
Above are exposure 0 residuals.
First run of MOSES_ALIGN_ORDERS was yesterday (log060523.163229). The routine was intentionally limited to registration of just the m=-1 order, for just the first two exposures. Summary of results:
CHISQ_BADNESS: chisq = 1744892.8 chisq_reduced = 0.85008617 imx = 24.1310 25.6349 2076.54 2075.01 imy = -3.62048 1019.20 -2.69529 1022.31 % COREGISTRATE: Alignment completed at Tue May 23 16:49:56 2006 ncalls = 555 .... CHISQ_BADNESS: chisq = 4656747.4 chisq_reduced = 2.2666494 imx = 26.4620 24.8723 2075.15 2075.08 imy = -3.50844 1018.37 -1.30873 1024.03 % COREGISTRATE: Alignment completed at Tue May 23 17:09:52 2006 ncalls = 701 moses_align_orders took 39.142551 minutes to run
IDL> print, alignment[0].minus.cpoints.x
24.1406 25.6308 2076.53 2074.99
IDL> print, alignment[0].minus.cpoints.y
-3.60734 1019.23 -2.75141 1022.29
IDL> print, alignment[1].minus.cpoints.x
26.4619 24.8725 2075.15 2075.08
IDL> print, alignment[1].minus.cpoints.y
-3.50831 1018.37 -1.30909 1024.03
IDL> print, alignment[0].minus.cpoints.x - alignment[1].minus.cpoints.x
-2.32132 0.758270 1.38257 -0.0922852
IDL> print, alignment[0].minus.cpoints.y - alignment[1].minus.cpoints.y
-0.0990317 0.858826 -1.44232 -1.74170
IDL> print, sqrt( (alignment[0].minus.cpoints.x - alignment[1].minus.cpoints.x)^2 + (alignment[0].minus.cpoints.y - alignment[1].minus.cpoints.y)^2 )
2.32343 1.14567 1.99794 1.74414
0.5 * 39min * 2 orders * 27 exposures = 17.6 hrs.
IDL> norm = median(cube_minus[*,*,1])/median(cube_plus[*,*,1])
IDL> print,norm
2.21052
IDL> foo1=cube_minus[*,*,1]-cube_plus[*,*,1]*norm
IDL> foo2=cube_minus[*,*,0]-cube_plus[*,*,0]*norm
IDL> write_jpeg, '20060524_residuals_exp0.jpg',bytscl(foo1)
% Loaded DLM: JPEG.
IDL> write_jpeg, '20060524_residuals_exp1.jpg',bytscl(foo2)
IDL> print, median(abs(foo1))
55.2104
IDL> print, median(abs(foo2))
14.5912
There is a silly mistake here, but it fortuitously cancels another silly mistake. See the notes from 2006-May-25, below.

Above are exposure 0 residuals.

Above are exposure 1 residuals.
% Compiled module: $MAIN$. % $MAIN$: Loading Level One MOSES data... Performing coregistration for exposure 0 % Compiled module: COREGISTRATE. % COREGISTRATE: Starting alignment at Wed May 24 15:26:55 2006 .... CHISQ_BADNESS: chisq = 1609928.1 chisq_reduced = 0.78340056 imx = 26.8515 25.6921 2074.50 2074.24 imy = -4.44583 1019.93 -0.765763 1022.88 % COREGISTRATE: Alignment completed at Wed May 24 15:35:21 2006 ncalls = 291 Performing coregistration for exposure 1 .... CHISQ_BADNESS: chisq = 4598330.9 chisq_reduced = 2.2376981 imx = 26.8659 25.5338 2074.61 2074.26 imy = -4.41270 1019.83 -0.887940 1022.93 % COREGISTRATE: Alignment completed at Wed May 24 15:42:41 2006 ncalls = 233 Performing coregistration for exposure 2 .... CHISQ_BADNESS: chisq = 9845535.5 chisq_reduced = 4.7908773 imx = 26.8523 25.4894 2074.61 2074.27 imy = -4.46986 1019.79 -0.737358 1023.08 % COREGISTRATE: Alignment completed at Wed May 24 15:50:12 2006 ncalls = 239 Performing coregistration for exposure 3 % COREGISTRATE: Starting alignment at Wed May 24 15:50:13 2006 CHISQ_BADNESS: chisq = 1.4621517e+10 chisq_reduced = 7118.7966 imx = 26.8527 25.4890 2074.61 2074.27 imy = -4.46948 1019.79 -0.737227 1023.08 .... CHISQ_BADNESS: chisq = 6.9963484e+09 chisq_reduced = 3406.7490 imx = 27.4948 25.2048 2077.28 2074.46 imy = -4.16954 1020.40 0.0537870 1023.45 % COREGISTRATE: Alignment completed at Wed May 24 16:01:13 2006 ncalls = 354 Performing coregistration for exposure 4 % COREGISTRATE: Starting alignment at Wed May 24 16:01:14 2006 CHISQ_BADNESS: chisq = 2.1569528e+11 chisq_reduced = 105881.32 imx = 27.4948 25.2048 2077.28 2074.46 imy = -4.16954 1020.40 0.0537912 1023.45 .... CHISQ_BADNESS: chisq = 1.9745229e+11 chisq_reduced = 96981.066 imx = 27.2258 25.5781 2077.50 2074.41 imy = -5.01160 1023.81 0.441277 1023.85 CHISQ_BADNESS: chisq = 1.9745229e+11 chisq_reduced = 96981.066 imx = 27.2258 25.5781 2077.50 2074.41 imy = -5.01160 1023.81 0.441277 1023.85 % COREGISTRATE: Alignment completed at Wed May 24 17:02:23 2006 ncalls = 2010 % COREGISTRATE: Amoeba failed to converge. % Execution halted at: COREGISTRATE 322
I've modified moses_align_orders.pro to save a snapshot (mosesAlignOrders_snap.sav) after every amoeba return. Then I can restore and run various things (like COREGISTRATE) by hand, with X11 graphical diagnostics.
Created automatic documentation system, docs, and reformatted the present notebook in HTML.
Looking at data from the last run made me realize several important things:
Sure enough, the error was in MOSES_PREP:
;Store in cube. cube_minus[*,*,j] = minus cube_zero[*,*,j] = plus cube_plus[*,*,j] = zero
The problem is in MOSES_PREP only: the movie programs were not affected. I have now fixed this.
Running prep again, moses_prep.log060525.171016.
Running MOSES_ALIGN_ORDERS again, moses_align_orders.log060525.172141.
Looking at residuals from the last run. The amoeba failed to converge on exposure 12. So exposures 1-11 should have the best alignments that MOSES_ALIGN_ORDERS can get, while 13-26 should be unmodified (as saved by MOSES_PREP).
I've written a simple procedure, EXAMINE_RESIDUALS, to step through the residuals and examine them visually. It turns out that none of the contents of cube_minus were modified by MOSES_ALIGN_ORDERS:
IDL> restore,'mosesAlignOrders_snap.sav
IDL> cube_minus_aligned = cube_minus
IDL> cube_zero_aligned = cube_zero
IDL> restore,'mosesLevelOne.sav
IDL> pmm, cube_minus[*,*,7] - cube_minus_aligned[*,*,7]
% Compiled module: PMM.
0.00000 0.00000
IDL> pmm, cube_minus[*,*,*] - cube_minus_aligned[*,*,*]
0.00000 0.00000
How strange! And by the way, cube_plus is also unmodified. I am running MOSES_ALIGN_ORDERS from the IDL command line to get some visual feedback. I have commented out set_plot, 'z' so that I'll be able to watch the residuals evolve.
OK, I've watched the residuals evolve through convergence of MOSES_ALIGN_ORDERS on exposure 0, using COREGISTRATE with CHISQ_BADNESS. The residuals look just the way I would expect, but somehow the modified m = -1 image does not get stuffed back into cube_minus.
Here is the call to COREGISTRATE from MOSES_ALIGN_ORDERS:
coregistrate, cube_zero[0:Nx-1,0:Ny-1,i], cube_minus[0:Nx-1, 0:Ny-1, i], $ 1, refx, refy, imx, imy, missing = missing_data, $ control_pts = control_pts, $ image2 = cube_minus[0:Nx-1, 0:Ny-1, i], $ Kx = alignment[i].minus.Kx, $ Ky = alignment[i].minus.Ky, $ /verboseThe image2 keyword should result in modification of cube_minus, shouldn't it? In a word, no. Have a look at this test program.
I have now fixed the bug in MOSES_ALIGN_ORDERS. Also restored the set_plot,'z' to allow it to run without an X display. Running again, moses_align_orders.log060530.134821
I have examined the residuals for the first bit of this run. Now they look right:
IDL> restore,'mosesAlignOrders_snap.sav' IDL> examine_residuals, cube_minus, cube_zero
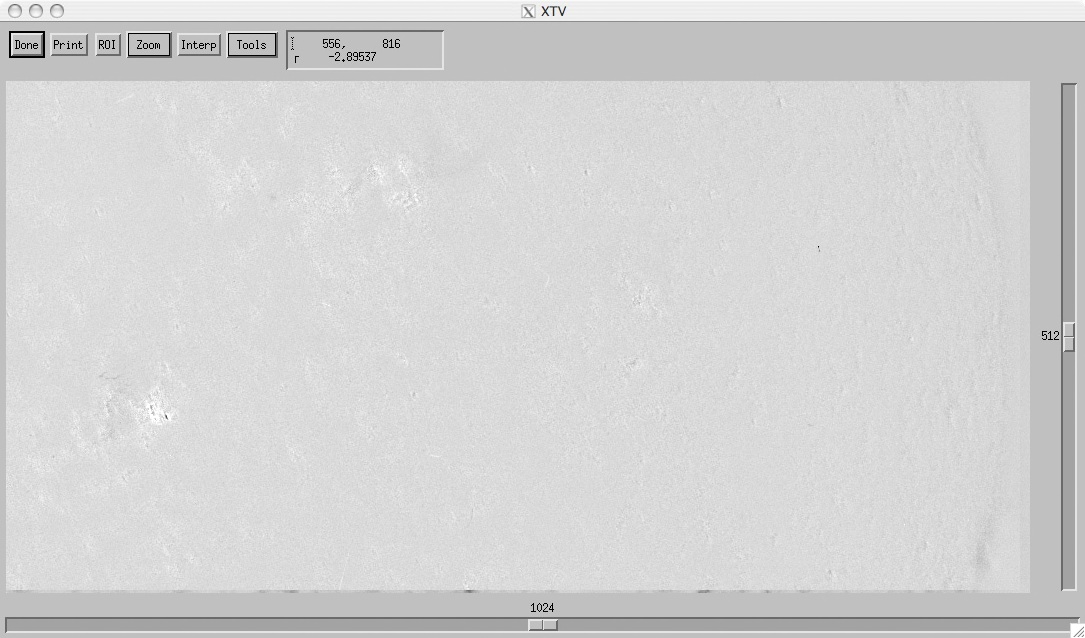 Above is the residual between orders m = -1 and m = 0 for exposure 0.
Above is the residual between orders m = -1 and m = 0 for exposure 0.
Some rationalization:
The idea behind this coregistration procedure is that the average doppler shift (over the whole image, or any large region) should be close to zero. This assumption may or may not be correct; however, it is common practice with many spectrometers to establish the 'line center' position based on an average over a quiet solar image. What we are doing is analogous. So I expect that if the global correlation is maximized as a function of alignment, then any remaining difference between any two orders will be attributable to:
The following questions are still not resolved:
Below is a detailed summary of the latest run, moses_align_orders.log060530.134821. It is useful to remember that the final alignment values for a particular exposure show up in the initial guess for the subsequent exposure.
Performing coregistration for exposure 0 % Compiled module: COREGISTRATE. % COREGISTRATE: Starting alignment at Tue May 30 13:53:29 2006 % Compiled module: AMOEBA. % Compiled module: POLYWARP. CHISQ_BADNESS: chisq = 8438225.3 chisq_reduced = 4.1061324 imx = 26.4000 25.0000 2075.00 2075.00 imy = -3.50000 1018.00 -1.30000 1024.00 .... CHISQ_BADNESS: chisq = 2156059.8 chisq_reduced = 1.0489861 imx = 21.1022 21.3940 2068.25 2068.36 imy = -5.81271 1017.92 -5.23799 1018.68 CHISQ_BADNESS: chisq = 2156056.5 chisq_reduced = 1.0489845 imx = 21.1017 21.3935 2068.25 2068.36 imy = -5.81227 1017.92 -5.23795 1018.68 % COREGISTRATE: Alignment completed at Tue May 30 14:05:13 2006 ncalls = 370 Performing coregistration for exposure 1 % COREGISTRATE: Starting alignment at Tue May 30 14:07:42 2006 CHISQ_BADNESS: chisq = 5435152.3 chisq_reduced = 2.6443604 imx = 21.1016 21.3944 2068.25 2068.36 imy = -5.81253 1017.92 -5.23825 1018.68 .... CHISQ_BADNESS: chisq = 5428321.6 chisq_reduced = 2.6410371 imx = 20.9810 21.5303 2068.30 2068.44 imy = -5.99142 1017.78 -5.12446 1018.74 % COREGISTRATE: Alignment completed at Tue May 30 14:15:18 2006 ncalls = 235 Performing coregistration for exposure 2 % COREGISTRATE: Starting alignment at Tue May 30 14:17:49 2006 CHISQ_BADNESS: chisq = 11390957. chisq_reduced = 5.5420333 imx = 20.9812 21.5304 2068.30 2068.44 imy = -5.99094 1017.78 -5.12439 1018.74 .... CHISQ_BADNESS: chisq = 11388261. chisq_reduced = 5.5408296 imx = 20.9575 21.5363 2068.34 2068.42 imy = -6.01806 1017.74 -5.02928 1018.82 CHISQ_BADNESS: chisq = 11388219. chisq_reduced = 5.5408093 imx = 20.9574 21.5364 2068.34 2068.42 imy = -6.01818 1017.74 -5.02916 1018.82 % COREGISTRATE: Alignment completed at Tue May 30 14:26:28 2006 ncalls = 267 Performing coregistration for exposure 3 % COREGISTRATE: Starting alignment at Tue May 30 14:29:05 2006 CHISQ_BADNESS: chisq = 3.5015171e+10 chisq_reduced = 17045.018 imx = 20.9573 21.5366 2068.34 2068.42 imy = -6.01847 1017.74 -5.02932 1018.82 .... CHISQ_BADNESS: chisq = 1.8526900e+10 chisq_reduced = 9035.9078 imx = 22.5858 23.9492 2069.63 2070.46 imy = -7.74742 1019.25 -5.62356 1017.42 CHISQ_BADNESS: chisq = 1.8526350e+10 chisq_reduced = 9035.6440 imx = 22.5859 23.9492 2069.63 2070.46 imy = -7.74775 1019.25 -5.62391 1017.42 % COREGISTRATE: Alignment completed at Tue May 30 14:36:53 2006 ncalls = 235 Performing coregistration for exposure 4 % COREGISTRATE: Starting alignment at Tue May 30 14:39:28 2006 CHISQ_BADNESS: chisq = 4.2707100e+11 chisq_reduced = 209790.01 imx = 22.5859 23.9492 2069.63 2070.46 imy = -7.74758 1019.25 -5.62372 1017.42 .... CHISQ_BADNESS: chisq = 3.5517067e+11 chisq_reduced = 174465.54 imx = 24.5128 24.0929 2069.38 2070.52 imy = -7.79466 1019.34 -5.48531 1019.22 CHISQ_BADNESS: chisq = 3.5517090e+11 chisq_reduced = 174465.65 imx = 24.5128 24.0929 2069.38 2070.52 imy = -7.79466 1019.34 -5.48531 1019.22 % COREGISTRATE: Alignment completed at Tue May 30 14:50:35 2006 ncalls = 349 Performing coregistration for exposure 5 % COREGISTRATE: Starting alignment at Tue May 30 14:53:19 2006 CHISQ_BADNESS: chisq = 2.6854113e+12 chisq_reduced = 1472806.8 imx = 24.5128 24.0929 2069.38 2070.52 imy = -7.79466 1019.34 -5.48531 1019.22 .... CHISQ_BADNESS: chisq = 2.2887503e+12 chisq_reduced = 1256278.8 imx = 25.2460 23.9374 2072.59 2071.33 imy = -6.94355 1018.75 -5.91531 1019.10 CHISQ_BADNESS: chisq = 2.2887498e+12 chisq_reduced = 1256278.5 imx = 25.2460 23.9374 2072.59 2071.33 imy = -6.94356 1018.75 -5.91531 1019.10 % COREGISTRATE: Alignment completed at Tue May 30 15:03:51 2006 ncalls = 336 Performing coregistration for exposure 6 % COREGISTRATE: Starting alignment at Tue May 30 15:06:34 2006 CHISQ_BADNESS: chisq = 5.3326029e+09 chisq_reduced = 2601.7253 imx = 25.2459 23.9374 2072.59 2071.33 imy = -6.94356 1018.75 -5.91531 1019.10 .... CHISQ_BADNESS: chisq = 2.0433226e+09 chisq_reduced = 996.29124 imx = 26.1697 26.2833 2072.23 2071.96 imy = -5.80089 1018.85 -5.13735 1017.84 CHISQ_BADNESS: chisq = 2.0429819e+09 chisq_reduced = 996.12514 imx = 26.1699 26.2829 2072.23 2071.96 imy = -5.80082 1018.85 -5.13719 1017.84 % COREGISTRATE: Alignment completed at Tue May 30 15:17:42 2006 ncalls = 347 Performing coregistration for exposure 7 % COREGISTRATE: Starting alignment at Tue May 30 15:20:11 2006 CHISQ_BADNESS: chisq = 3.2378295e+09 chisq_reduced = 1578.6982 imx = 26.1699 26.2830 2072.23 2071.96 imy = -5.80085 1018.85 -5.13725 1017.84 .... CHISQ_BADNESS: chisq = 2.5994588e+09 chisq_reduced = 1266.0055 imx = 26.1625 26.2760 2072.30 2071.96 imy = -3.83107 1018.80 -4.91849 1018.04 CHISQ_BADNESS: chisq = 2.5992067e+09 chisq_reduced = 1265.8828 imx = 26.1625 26.2760 2072.30 2071.96 imy = -3.83107 1018.80 -4.91849 1018.05 % COREGISTRATE: Alignment completed at Tue May 30 15:30:20 2006 ncalls = 317 Performing coregistration for exposure 8 % COREGISTRATE: Starting alignment at Tue May 30 15:33:21 2006 CHISQ_BADNESS: chisq = 2.9760573e+09 chisq_reduced = 1449.4183 imx = 26.1625 26.2760 2072.30 2071.96 imy = -3.83107 1018.80 -4.91849 1018.05 .... CHISQ_BADNESS: chisq = 2.9563196e+09 chisq_reduced = 1439.8659 imx = 26.1202 26.3516 2072.34 2071.87 imy = -3.96789 1018.85 -4.74246 1018.39 CHISQ_BADNESS: chisq = 2.9553041e+09 chisq_reduced = 1439.3706 imx = 26.1202 26.3515 2072.34 2071.87 imy = -3.96784 1018.85 -4.74266 1018.39 % COREGISTRATE: Alignment completed at Tue May 30 15:42:10 2006 ncalls = 273 Performing coregistration for exposure 9 % COREGISTRATE: Starting alignment at Tue May 30 15:44:44 2006 CHISQ_BADNESS: chisq = 3.0861590e+09 chisq_reduced = 1503.0905 imx = 26.1202 26.3516 2072.34 2071.87 imy = -3.96796 1018.85 -4.74265 1018.39 .... CHISQ_BADNESS: chisq = 2.1939274e+09 chisq_reduced = 1067.3553 imx = 25.9750 26.5327 2072.51 2072.03 imy = -2.26547 1018.44 -4.28019 1018.61 CHISQ_BADNESS: chisq = 2.1938828e+09 chisq_reduced = 1067.3335 imx = 25.9750 26.5327 2072.51 2072.03 imy = -2.26547 1018.44 -4.28020 1018.61 % COREGISTRATE: Alignment completed at Tue May 30 15:55:11 2006 ncalls = 319 Performing coregistration for exposure 10 % COREGISTRATE: Starting alignment at Tue May 30 15:57:48 2006 CHISQ_BADNESS: chisq = 2.4845427e+09 chisq_reduced = 1208.7415 imx = 25.9750 26.5327 2072.51 2072.03 imy = -2.26547 1018.44 -4.28019 1018.61 .... CHISQ_BADNESS: chisq = 1.5098930e+09 chisq_reduced = 733.59415 imx = 26.3855 26.1889 2072.22 2071.90 imy = 0.0127509 1018.70 -4.01597 1018.86 CHISQ_BADNESS: chisq = 1.5098923e+09 chisq_reduced = 733.59378 imx = 26.3855 26.1888 2072.22 2071.90 imy = 0.0127462 1018.70 -4.01598 1018.86 % COREGISTRATE: Alignment completed at Tue May 30 16:08:10 2006 ncalls = 324 Performing coregistration for exposure 11 % COREGISTRATE: Starting alignment at Tue May 30 16:10:39 2006 CHISQ_BADNESS: chisq = 2.0484360e+09 chisq_reduced = 995.22948 imx = 26.3855 26.1888 2072.22 2071.90 imy = 0.0127432 1018.70 -4.01597 1018.86 .... CHISQ_BADNESS: chisq = 1.1040684e+09 chisq_reduced = 536.13773 imx = 26.3855 26.1888 2072.22 2071.90 imy = 0.0793048 1021.61 -3.01601 1018.86 CHISQ_BADNESS: chisq = 1.1040678e+09 chisq_reduced = 536.13742 imx = 26.3855 26.1888 2072.22 2071.90 imy = 0.0793013 1021.61 -3.01599 1018.86 % COREGISTRATE: Alignment completed at Tue May 30 16:17:06 2006 ncalls = 200 Performing coregistration for exposure 12 % COREGISTRATE: Starting alignment at Tue May 30 16:19:32 2006 CHISQ_BADNESS: chisq = 2.0316974e+09 chisq_reduced = 986.60952 imx = 26.3855 26.1888 2072.22 2071.90 imy = 0.0793936 1021.61 -3.01597 1018.86 ....
The log file has become too big to grok by inspection. Am developing a procedure to visualize the consistency (or lack thereof) in the alignments: ALIGN_SUMMARY. Alignment summary image align_summary2006May31113325.jpg, produced for the latest run, is below:
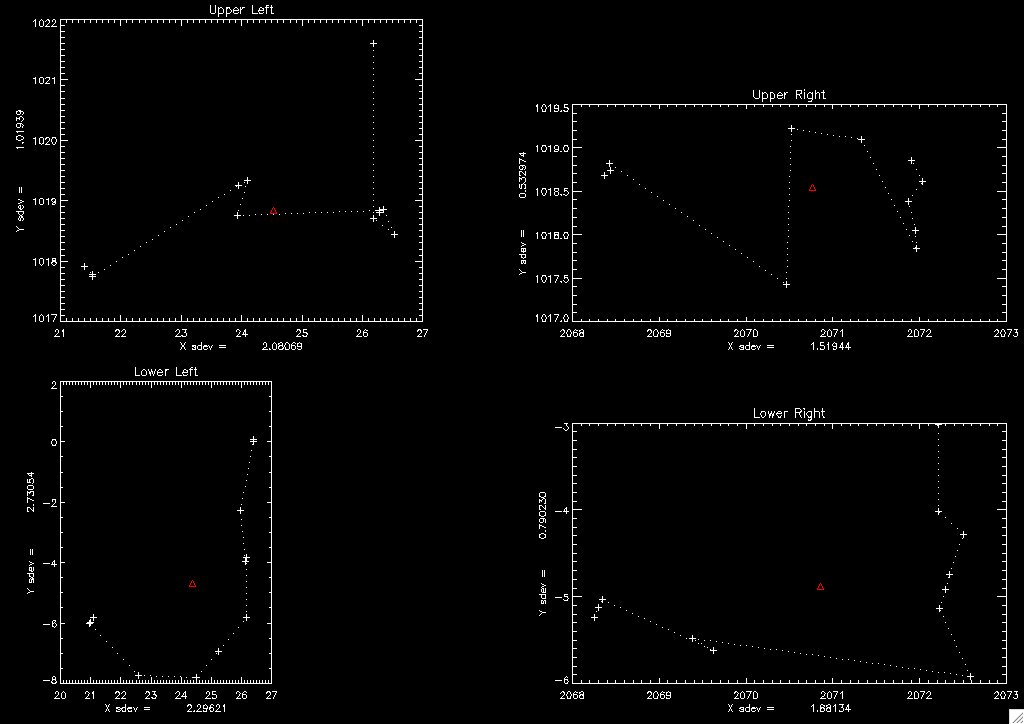
Last night's summary image shows two symptoms of disease:
Ideas (in the ordeer thay occurred to me, not in order of priority):
While checking 1 and 2, I added a few more comments to the code but did not change anything functionally.
Wrote SAVESCREEN to make documentation more convenient. This is the screen image saving code from ALIGN_SUMMARY.
Using EXAMINE_RESIDUALS, I see plausible alignments for exposures 0,1,2. In exposures 3 and 6, the minus order is apparently shifted left relative to zero. The apparent bipolar features are consistent throughout the residuals image.
It is difficult to examine the longer exposures, because data that should be marked missing in the image2 keyword output of COREGISTRATE isn't. This is the old POLY_2D bug (see modification notes in COREGISTRATE). I have now carried through the fix that was used for the badness procedures to the image2 keyword output. Time to run the alignment again, so that the data cubes are modified accordingly.
Re-running the alignment: moses_align_orders.log060531.134357. Oops, had to fix a bug in my mods to COREGISTRATE. Re-running. moses_align_orders.log060531.140206.