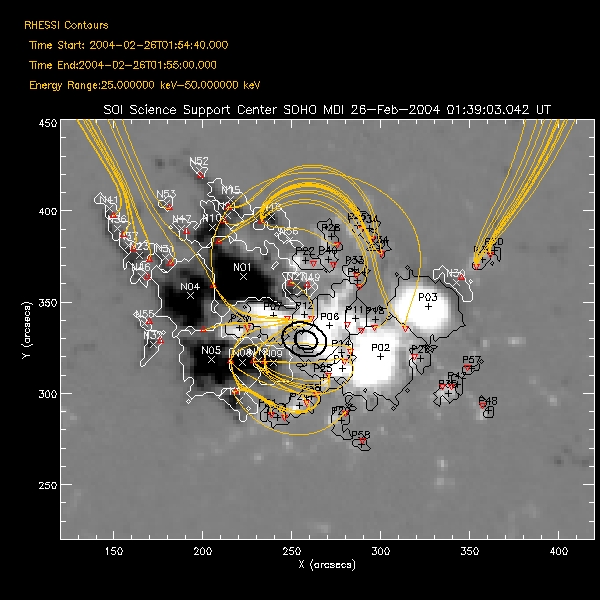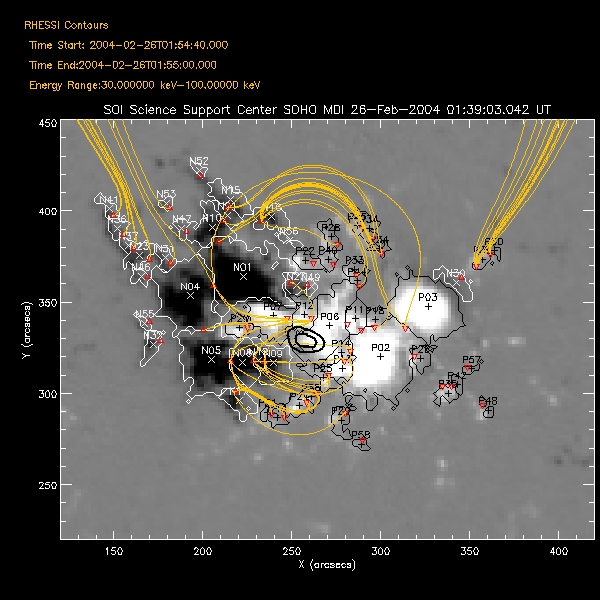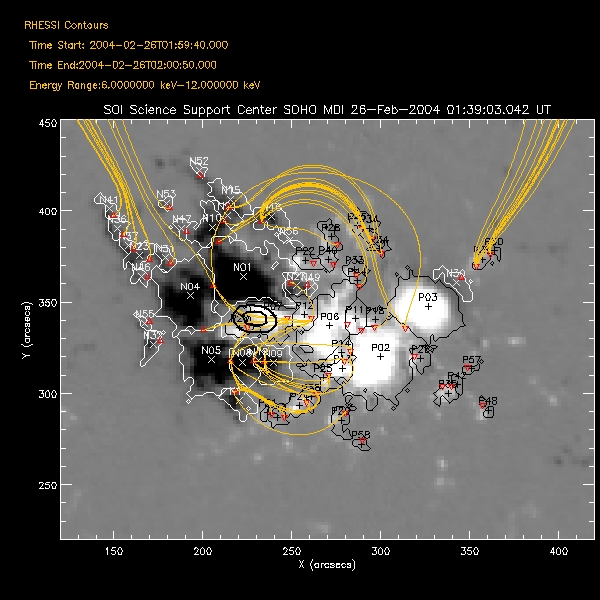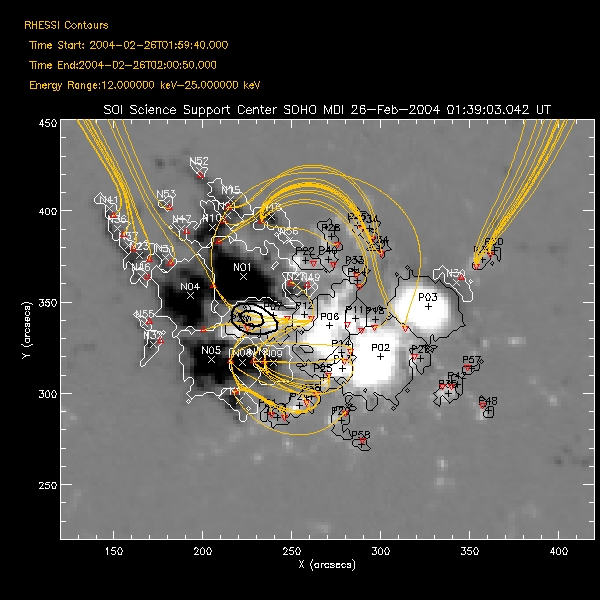
| Saving
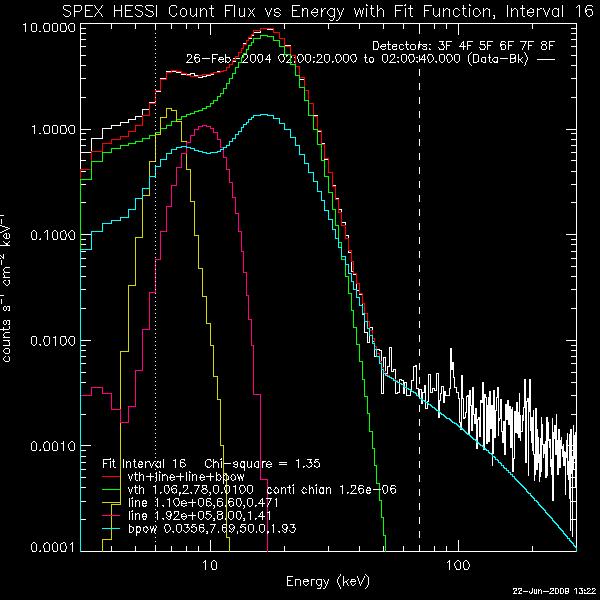
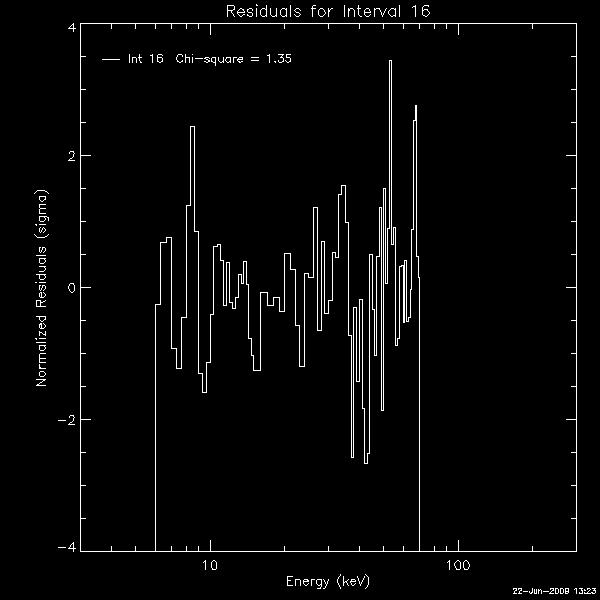
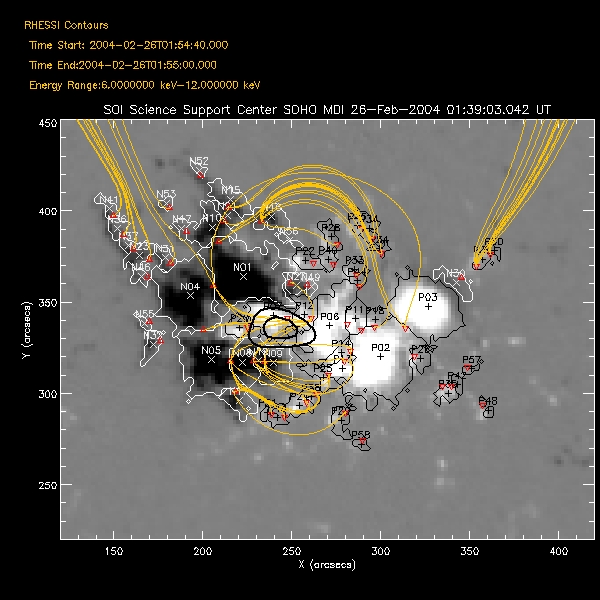
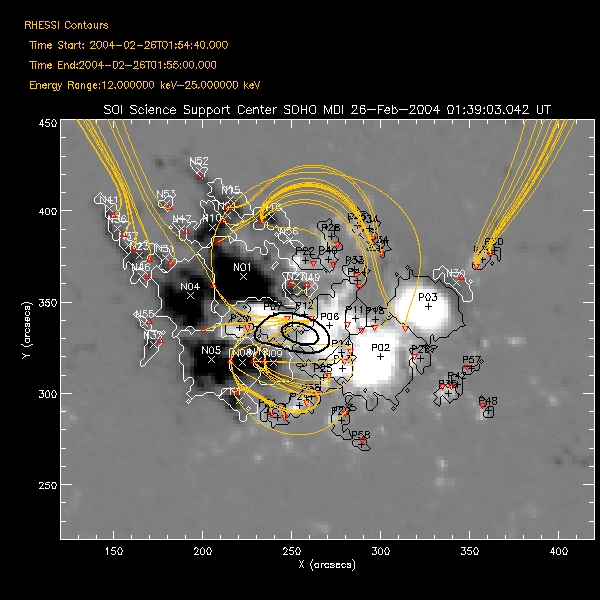
and Restoring OSPEX Session The writescript method saves an OSPEX session. Internally, there are two steps to saving an OSPEX session: 1. Storing the current value of all control parameters that are not at their current value. This is accomplished by writing a procedure script that creates an OSPEX object and issues the SET command for each of those changed parameters. 2. Saving the result of fitting that has been done. This is accomplished by writing all of the fit results parameters (all info parameters starting with spex_summ) into an output file (FITS format). (See the section on Saving and Restoring OSPEX Results to see how to do this step by itself.) In the OSPEX GUI, use the buttons under File to 'Write Script'. Additional buttons let you choose whether to write a fit results file as well as the script file. A widget will appear to allow you to choose the names for the script file and the fit results file. The name of the procedure will be the name of the script file you select. At the command line, type (see the writescript description for calling arguments): o -> writescript To run the script to restore the session, type script_name, obj=obj If obj is an existing OSPEX object, then the procedure will set the parameters in that object to the values they had when you wrote the script (Note - parameters not mentioned in the script will be unchanged, they will not be initialized). If not, the procedure will create a new OSPEX object and set the parameters/values. Depending on how you wrote the script, it may start the GUI, and and it may restore the fit results. In addition, there is a button on the GUI under File to 'Set Parameters from Script' to allow you to set parameters in an existing object according to the script file (either initializing all parameters to defaults first or not). At the command line, type: o->runscript or o->runscript, /init If o is an existing object, then script_name,obj=o is equivalent to o->runscript. Saving and Restoring OSPEX Results The fit results are saved via the OSPEX savefit method which writes all of the fit results parameters (all info parameters starting with spex_summ) into a file. They are restored into the OSPEX object via the restorefit method which reads the file and uses the SET method to set the spex_summ parameters into the object. They are restored to your IDL session via the spex_read_fit_results routine. Two types of output file are supported: * FITS - This is the preferred format. FITS is a standard format and can be read by FITS readers in many languages; it does not require IDL or SSW. Another advantage of the FITS format is that context information (including most of the OSPEX control parameter values) is included in the FITS file. The disadvantage is that the files are bigger than the geny files. * Save (geny) - No longer supported as of 7-May-2006. The geny files are a refinement of the basic IDL save files, written and read by the SSW routines savegenx and restgenx. They are smaller than the FITS files, but the format is specific to IDL/SSW. In the OSPEX GUI, use the buttons under File to 'Save Fit Results' and 'Import Fit Results'. At the command line, type the following to save the fit results or restore fit results into the OSPEX object: o -> savefit o -> restorefit Note that it is possible to include restoring the fit results as part of a script that restores an OSPEX session (see section on Saving and Restoring OSPEX Session) To restore the fit results structure directly (without creating an OSPEX object) from either a FITS or geny file, use the spex_read_fit_results procedure. For example, fit_results = spex_read_fit_results('ospex_results.fits') (Geny files can also be read in directly using the restgenx command: restgenx, file='ospex_results.geny', fit_results ) The structure containing all of the fit results will be returned in the fit_results variable. Look at the parameters starting with spex_summ in the OSPEX Info Parameter Table for the meaning of each tag in the structure. |